Research
Research Assistant
Abstract
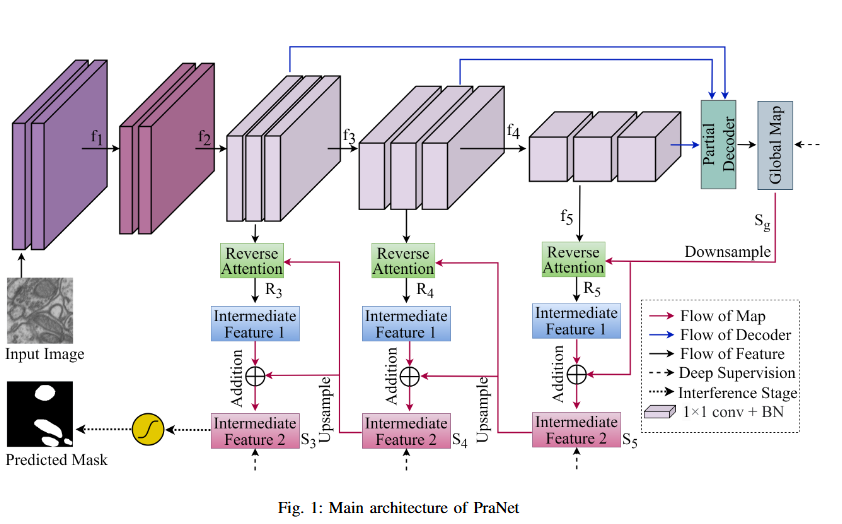
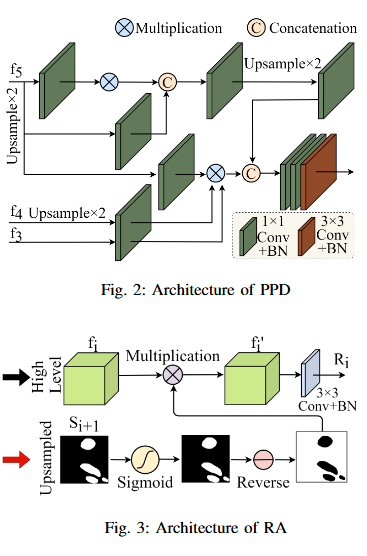
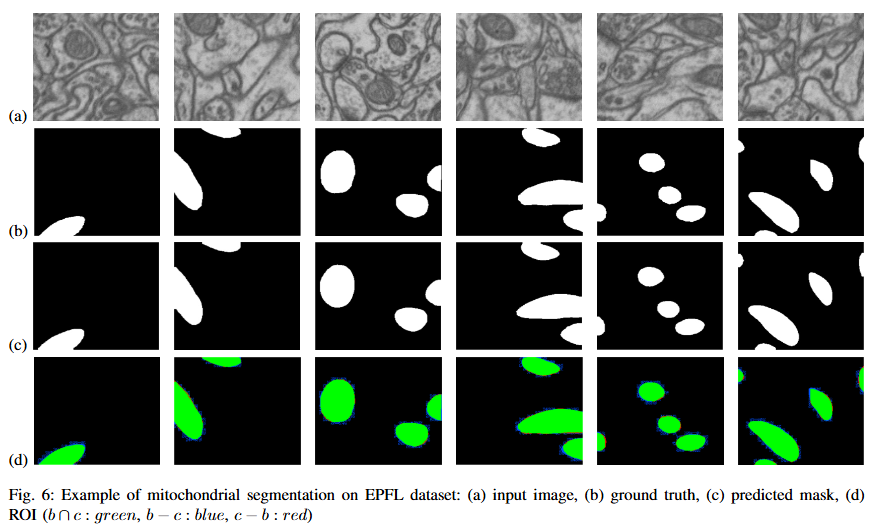
Mitochondrial segmentation in microscopic images is a complex task hindered by variations in mitochondrial size, color, texture, and their often ambiguous borders with surrounding tissue. To overcome these obstacles, we adapted Parallel Reverse Attention Network (Pranet), originally developed for precise polyp detection, to the domain of mitochondrial segmentation. Pranet employs Parallel Partial Decoder (PPD) to integrate multi-level features, producing a high-level context map that guides subsequent processing. The model further enhances segmentation precision through a Reverse Attention (RA) module, which adeptly identifies object boundaries by iteratively refining region-boundary interactions. Our approach was thoroughly evaluated on the EPFL dataset, yielding substantial improvements in segmentation accuracy. Additionally, Pranet demonstrated robust generalization and real-time performance, underscoring its suitability for comprehensive microscopic image analysis.
BibTeX
Click to copy@INPROCEEDINGS{11013970,
author={Alam, Ayat Subah and Rhythm, Faiaz Hasanuzzaman and Ovi, Tareque Bashar and Bashree, Nomaiya and Nyeem, Hussain and Wahed, Md Abdul},
booktitle={2025 International Conference on Electrical, Computer and Communication Engineering (ECCE)},
title={Utilizing Reverse Attention for Enhanced Mitochondria Segmentation in Microscopic Images},
year={2025},
volume={},
number={},
pages={1-6},
keywords={Image segmentation;Adaptation models;Mitochondria;Accuracy;Image color analysis;Microscopy;Computer architecture;Transformers;Real-time systems;Biomedical imaging;deep learning;medical image segmentation;contextual information;mitochondria segmentation;electron-microscopy images;reverse attention},
doi={10.1109/ECCE64574.2025.11013970}}

Abstract
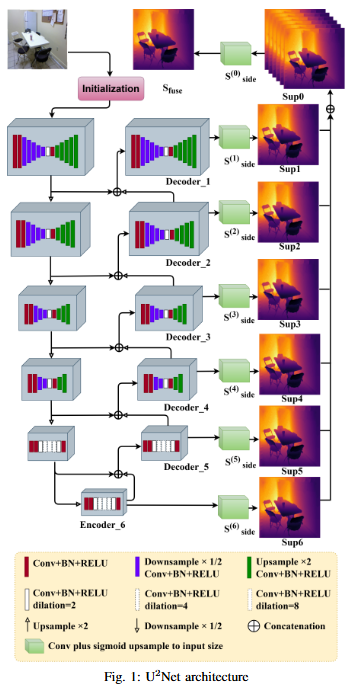
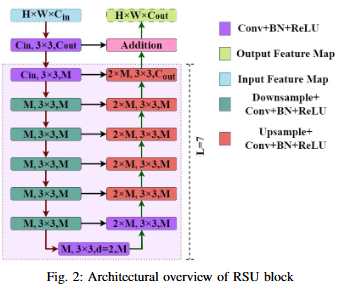
Monocular depth estimation (MDE) has evolved into a vital research focus in visual computing and autonomous robotic systems. While recent deep learning approaches have advanced the field, existing methods often struggle to jointly incorporate global context and fine-grained local details, and may be hampered by complex, computationally expensive network architectures. To address these challenges, we employ the U2Net framework, which incorporates Residual U-Blocks designed to adapt the area of reception and capture multi-scale contextual data. By nesting U-shaped structures, U2Net effectively increases feature extraction depth while maintaining computational efficiency, thus offering a more balanced and scalable solution for MDE. Our proposed model, developed using the NYUDepth V2 dataset, exceeds contemporary methods in critical assessment measures, including RMSE, RMSE(log), and Absolute Relative Error (Abs Rel). These improved results highlight the robustness and precision of proposed model or a range of practical applications in robotics, augmented reality, and other domains requiring real-time scene geometry understanding.
BibTeX
Click to copy@INPROCEEDINGS{11013114,
author={Rhythm, Faiaz Hasanuzzaman and Bashar Ovi, Tareque and Bashree, Nomaiya and Islam Ratul, Md. Raisul and Nyeem, Hussain and Wahed, Md Abdul},
booktitle={2025 International Conference on Electrical, Computer and Communication Engineering (ECCE)},
title={Optimizing Monocular Depth Estimation through Bi-Level Nested Architecture Integration},
year={2025},
volume={},
number={},
pages={1-6},
keywords={Deep learning;Adaptation models;Visualization;Depth measurement;Computational modeling;Reliability engineering;Robustness;Real-time systems;Computational efficiency;Robots;deep learning;depth estimation;contextual and local information;residual u-block;nested u-structure},
doi={10.1109/ECCE64574.2025.11013114}}
Abstract
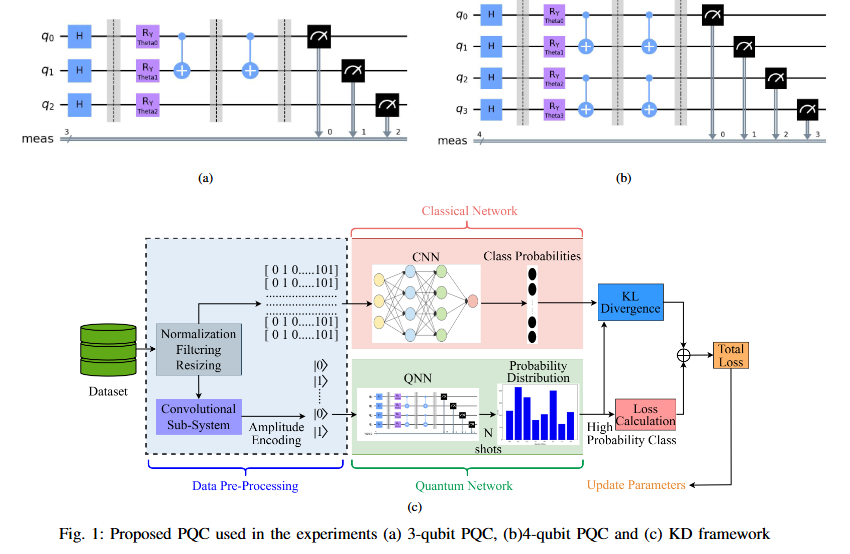
While Quantum Neural Networks (QNNs) offer the potential for resource-efficient image classification, practical implementations are hindered by qubit instability and finite availability, causing them to often underperform classical counterparts. This performance disparity constitutes a critical research gap. We propose a novel hybrid classical-quantum knowledge distillation approach to bridge this gap. Our method utilizes a pre-trained classical CNN (AlexNet) as a teacher to guide the training of a quantum student model by minimizing the Kullback-Leibler (KL) divergence (DKL) between their output distributions. The quantum student model employs an architecture designed to facilitate intricate feature interactions using iterated CNOT gates. This technique substantially improves quantum model accuracy, demonstrating gains of 0.34% (6-class, 3 qubits) and 0.28% (8-class, 3 qubits) on the MNIST dataset, and 0.74% (8-class, 4 qubits). Our contribution lies in operationalizing a knowledge distillation paradigm for applied Quantum Machine Learning (QML), enabling the effective transfer of robust knowledge from established classical architectures to resource-constrained quantum models, thereby enhancing faster convergence and accelerating practical QML deployment.
BibTeX
Click to copy@INPROCEEDINGS{11159974,
author={Ovi, Tareque Bashar and Bashree, Nomaiya and Rhythm, Faiaz Hasanuzzaman and Chowdhury, Disha and Nyeem, Hussain and Wahed, Md Abdul},
booktitle={2025 2nd International Conference on Next-Generation Computing, IoT and Machine Learning (NCIM)},
title={Bridging Classical and Quantum Models via Attention-Guided Feature Distillation},
year={2025},
volume={},
number={},
pages={1-6},
keywords={Training;Accuracy;Qubit;Transfer learning;Neural networks;Computer architecture;Logic gates;Quantum networks;Next generation networking;Image classification;Deep learning;knowledge distillation;amplitude encoding;transfer learning;QML},
doi={10.1109/NCIM65934.2025.11159974}}
Abstract
This paper presents YESnet, a novel framework integrating the YOLOv11 object detector with the Segment Anything Model 2 (SAM-2) for memory-efficient skin lesion segmentation. The proposed architecture introduces: (1) An anchor-free YOLOv11 detection head that eliminates predefined bounding box priors while maintaining detection accuracy, (2) A memory-optimized SAM-2 variant that preserves boundary delineation capabilities through compressed feature retention, and (3) A lightweight co-processing pipeline that coordinates detection and segmentation stages without computational redundancy. Evaluated on the HAM10000 dataset, YESnet achieves competitive performance in lesion segmentation (96.00% accuracy, 92.38% Dice) while demonstrating efficient memory utilization. The framework's design enables effective lesion boundary capture with minimal parameter overhead, making it particularly suitable for resource-constrained clinical deployment scenarios.
BibTeX
Click to copy@INPROCEEDINGS{11171738,
author={Bashree, Nomaiya and Ovi, Tareque Bashar and Zahid, Sadia Binte and Rhythm, Faiaz Hasanuzzaman and Nyeem, Hussain and Abdul, Md},
booktitle={2025 International Conference on Quantum Photonics, Artificial Intelligence, and Networking (QPAIN)},
title={YESnet: YOLOv11 Enabled SAM-2 Framework for Memory-Efficient Skin Lesion Segmentation},
year={2025},
volume={},
number={},
pages={1-5},
keywords={Image segmentation;Accuracy;Image analysis;Pipelines;Redundancy;Memory management;Detectors;Skin;Lesions;Photonics;Deep learning;skin lesion segmentation;foundational model;nano YOLOv11 architecture;SAM-2},
doi={10.1109/QPAIN66474.2025.11171738}}

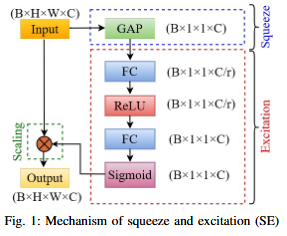
Abstract
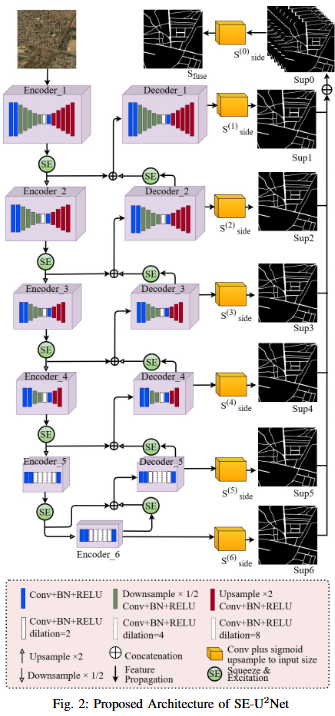
Road extraction from very high-resolution satellite imagery (VHR) is necessary for remote sensing applications like city management, and navigation. In this study, we introduce SE-U2Net, a novel deep learning model for precise road extraction. Our proposed model integrates the Squeeze and Excitation (SE) network with the U2 Net architecture for enhanced selective channel refinement and better contextual information capture. SE-U2Net employs Residual U-blocks (RSU), which combine receptive fields of different sizes, enabling the model to capture multiscale contextual data efficiently. The baseline U2Net has 75.06 GFLOPs and 37.51 GMacs, while SE-U2Net maintains this efficiency with 75.07 GFLOPs and 37.51 GMacs, despite its enhanced representational capacity through SE blocks. Emperical outcomes on the DeepGlobe dataset demonstrate that SE-U2Net outperforms the base U2Net model and other state-of-the-art models, achieving superior performance in terms of Dice coefficient, mean Intersection over Union (mIoU), precision, and recall. These results underscore SE-U2Net's potential as a robust tool for high-accuracy road segmentation from VHR satellite imagery.
BibTeX
Click to copy@INPROCEEDINGS{11171628,
author={Rhythm, Faiaz Hasanuzzaman and Ovi, Tareque Bashar and Bashree, Nomaiya and Nyeem, Hussain and Wahed, Md Abdul},
booktitle={2025 International Conference on Quantum Photonics, Artificial Intelligence, and Networking (QPAIN)},
title={Enhancing U2Net for Precise Road Extraction from Satellite Images via Channel Refinement},
year={2025},
volume={},
number={},
pages={1-6},
keywords={Deep learning;Image segmentation;Adaptation models;Roads;Computational modeling;Urban areas;Computer architecture;Satellite images;Computational efficiency;Context modeling;Road extraction;Segmentation;Skip connection;Squeeze-excitation;High-resolution satellite image},
doi={10.1109/QPAIN66474.2025.11171628}}
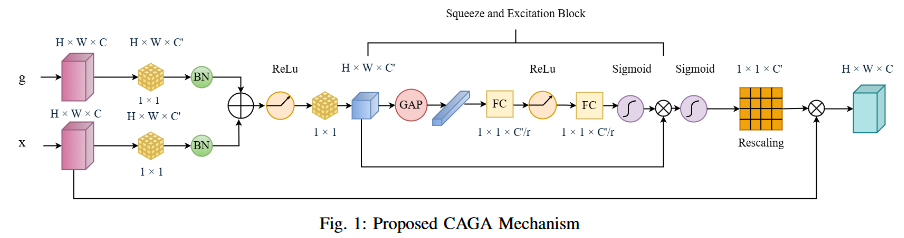
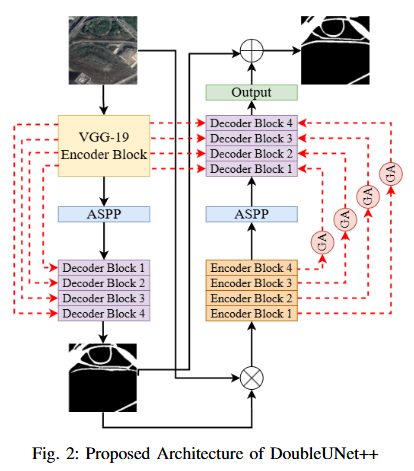
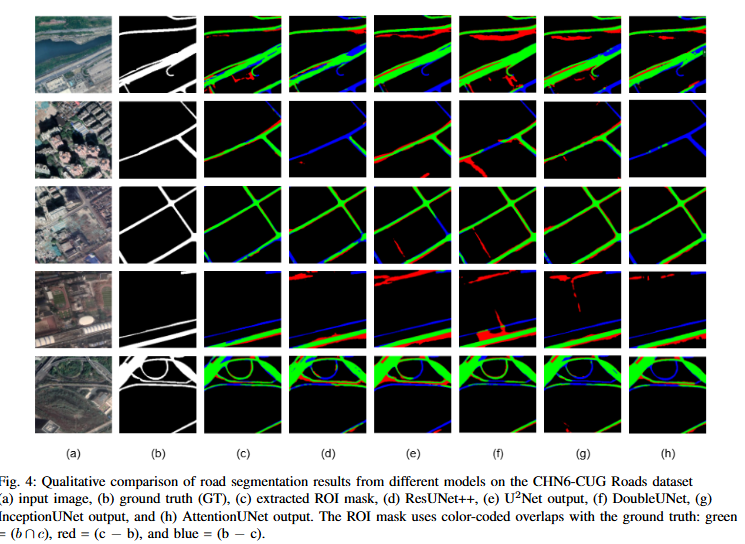
Abstract
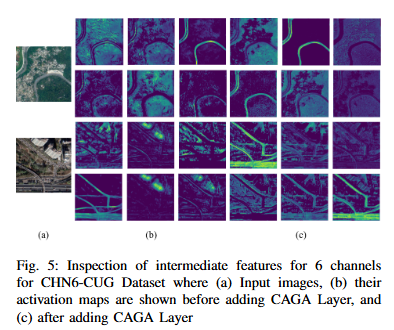
Road extraction from satellite imagery faces critical challenges in capturing contextual spatial relationships and mitigating background interference. We introduce DoubleUNet++, which improves the DoubleUNet architecture with two main enhancements: (1) A channel-aware gated attention (CAGA) mechanism in the skip connections of the second U-Net, dynamically weighting channel features via decoder gating signals, and (2) Squeeze-and-Excitation layers in the attention path for adaptive channel-wise feature recalibration. Evaluated on the CHN6-CUG dataset (4,511 annotated images), our model achieves state-of-the-art performance with 78.52 Dice score and 64.94 mIoU - representing 3.32 % and 3.74 % absolute gains over baseline DoubleUNet. Despite these improvements, DoubleUNet++ maintains similar computational complexity (109.33 GFLOPs, 54.64 GMacs) compared to the baseline (107.41 GFLOPs, 53.68 GMacs). The architecture maintains comparable complexity (29.5M parameters) while significantly improving precision (81.45% vs 73.78%) through refined spatial focus on roadrelevant regions. Qualitative analysis demonstrates the CAGA mechanism's effectiveness in reducing misclassification from nonroad objects such as vegetation and buildings while preserving road topology. These advancements establish DoubleUNet++ as a robust solution for large-scale road network mapping without requiring additional annotated data.
BibTeX
Click to copy@INPROCEEDINGS{11172088,
author={Rhythm, Faiaz Hasanuzzaman and Bashree, Nomaiya and Ovi, Tareque Bashar and Nyeem, Hussain and Wahed, Md Abdul},
booktitle={2025 International Conference on Quantum Photonics, Artificial Intelligence, and Networking (QPAIN)},
title={DoubleUNet++: Channel-Aware Gated Attention for Road Extraction in Satellite Imagery},
year={2025},
volume={},
number={},
pages={1-6},
keywords={Image segmentation;Roads;Vegetation mapping;Computer architecture;Logic gates;Feature extraction;Data models;Spatial databases;Satellite images;Topology;Deep learning;Road extraction;Cascaded model;Segmentation;Gated attention;Skip connection;Squeeze-excitation;High-resolution satellite image},
doi={10.1109/QPAIN66474.2025.11172088}}
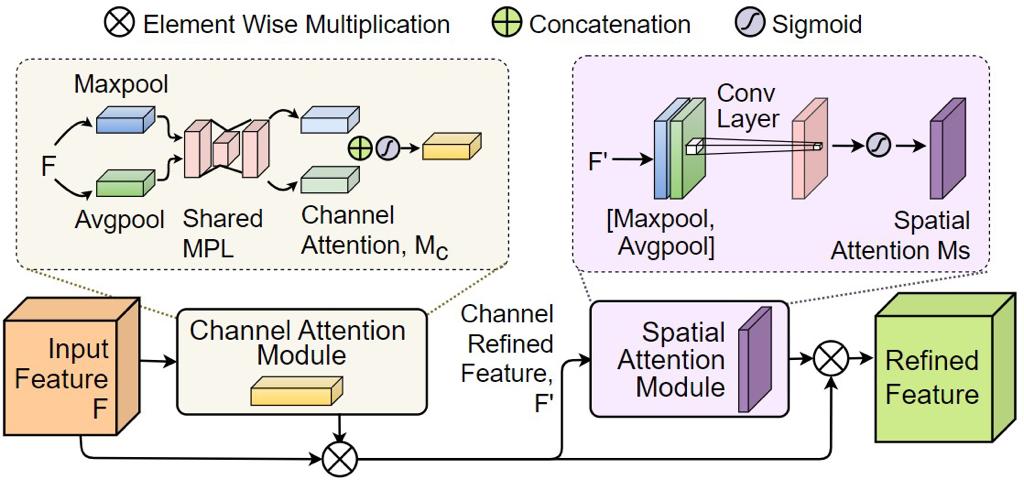
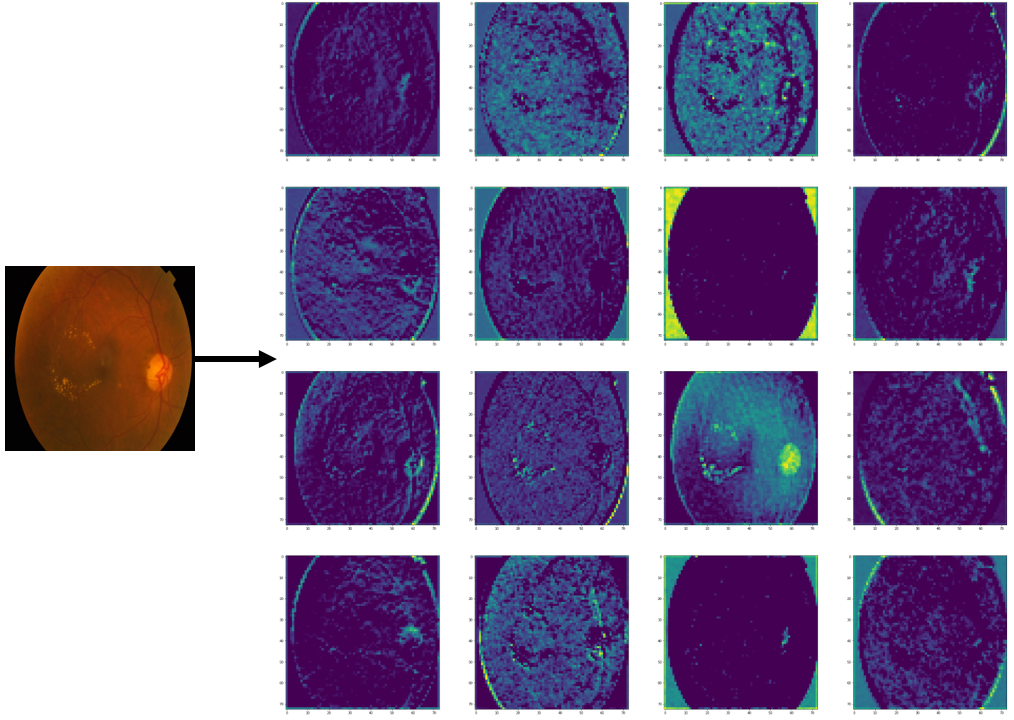
Abstract
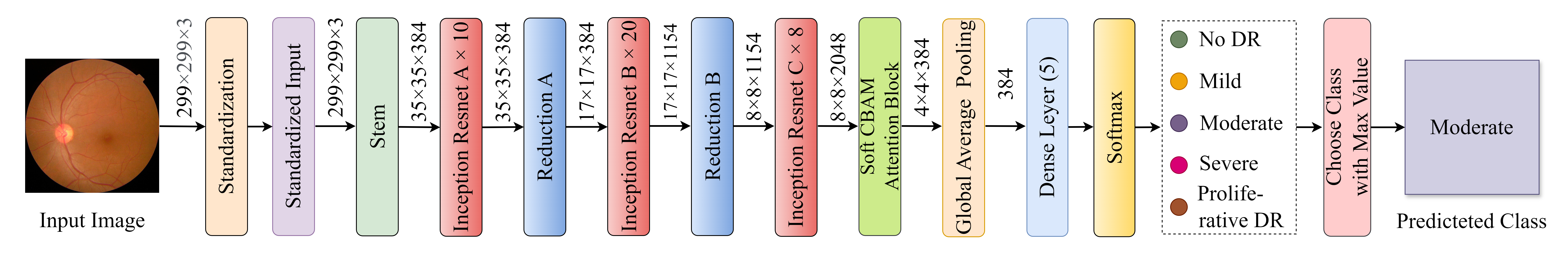
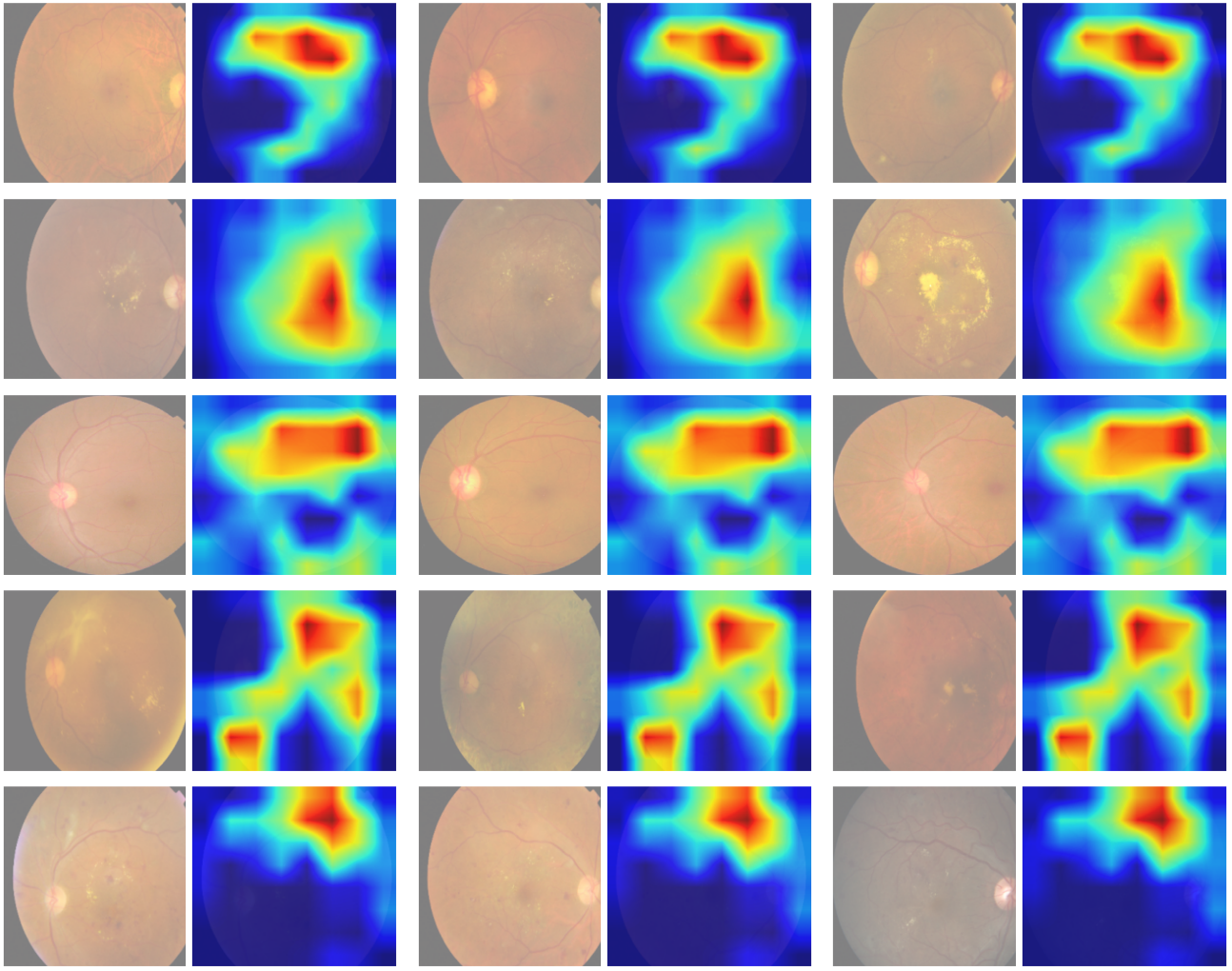
Diabetic retinopathy (DR) poses a serious threat to vision, emphasising the need for early detection. Manual analysis of fundus images, though common, is error-prone and time-intensive. Existing automated diagnostic methods lack precision, particularly in the early stages of DR. This paper introduces the Soft Convolutional Block Attention Module-based Network (Soft-CBAMNet), a deep learning network designed for severity detection, which features Soft-CBAM attention to capture complex features from fundus images. The proposed network integrates both the convolutional block attention module (CBAM) and the soft-attention components, ensuring simultaneous processing of input features. Following this, attention maps undergo a max-pooling operation, and refined features are concatenated before passing through a dropout layer with a dropout rate of 50%. Experimental results on the APTOS dataset demonstrate the superior performance of Soft-CBAMNet, achieving an accuracy of 85.4% in multiclass DR grading. The proposed architecture has shown strong robustness and general feature learning capability, achieving a mean AUC of 0.81 on the IDRID dataset. Soft-CBAMNet's dynamic feature extraction capability across all classes is further justified by the inspection of intermediate feature maps. The model excels in identifying all stages of DR with increased precision, surpassing contemporary approaches. Soft-CBAMNet presents a significant advancement in DR diagnosis, offering improved accuracy and efficiency for timely intervention.
BibTeX
Click to copy@article{https://doi.org/10.1002/ima.23175,
author = {Ovi, Tareque Bashar and Bashree, Nomaiya and Nyeem, Hussain and Wahed, Md Abdul and Rhythm, Faiaz Hasanuzzaman and Alam, Ayat Subah},
title = {A Novel Dual Attention Approach for DNN Based Automated Diabetic Retinopathy Grading},
journal = {International Journal of Imaging Systems and Technology},
volume = {34},
number = {5},
pages = {e23175},
doi = {https://doi.org/10.1002/ima.23175},
url = {https://onlinelibrary.wiley.com/doi/abs/10.1002/ima.23175},
eprint = {https://onlinelibrary.wiley.com/doi/pdf/10.1002/ima.23175},
note = {e23175 IMA-24-504.R1},
abstract = {ABSTRACT Diabetic retinopathy (DR) poses a serious threat to vision, emphasising the need for early detection. Manual analysis of fundus images, though common, is error-prone and time-intensive. Existing automated diagnostic methods lack precision, particularly in the early stages of DR. This paper introduces the Soft Convolutional Block Attention Module-based Network (Soft-CBAMNet), a deep learning network designed for severity detection, which features Soft-CBAM attention to capture complex features from fundus images. The proposed network integrates both the convolutional block attention module (CBAM) and the soft-attention components, ensuring simultaneous processing of input features. Following this, attention maps undergo a max-pooling operation, and refined features are concatenated before passing through a dropout layer with a dropout rate of 50\%. Experimental results on the APTOS dataset demonstrate the superior performance of Soft-CBAMNet, achieving an accuracy of 85.4\% in multiclass DR grading. The proposed architecture has shown strong robustness and general feature learning capability, achieving a mean AUC of 0.81 on the IDRID dataset. Soft-CBAMNet's dynamic feature extraction capability across all classes is further justified by the inspection of intermediate feature maps. The model excels in identifying all stages of DR with increased precision, surpassing contemporary approaches. Soft-CBAMNet presents a significant advancement in DR diagnosis, offering improved accuracy and efficiency for timely intervention.},
year = {2024}
}
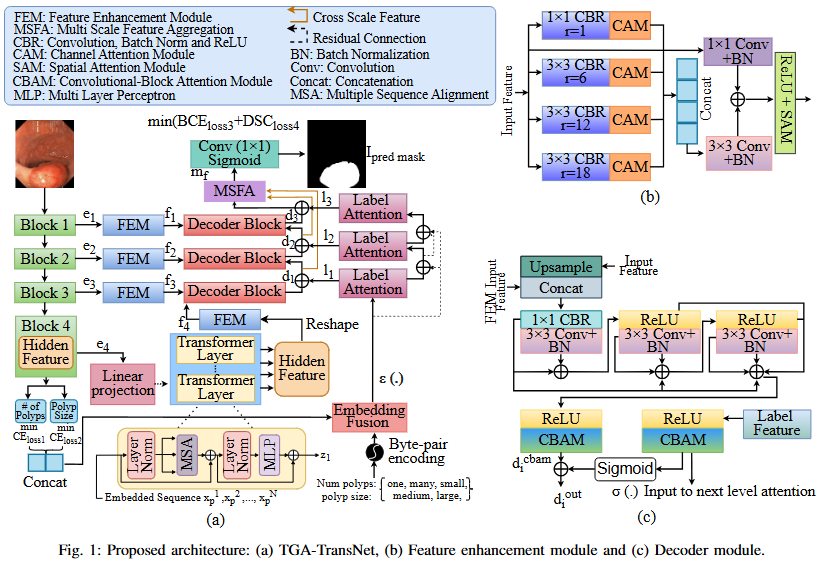
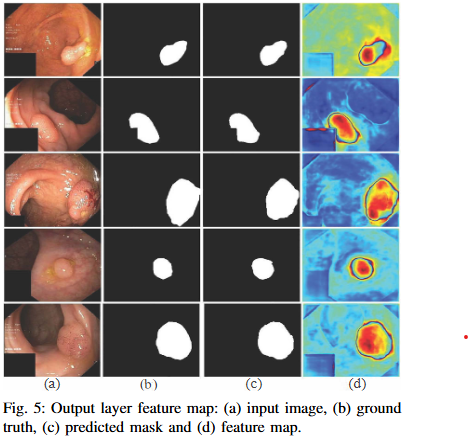
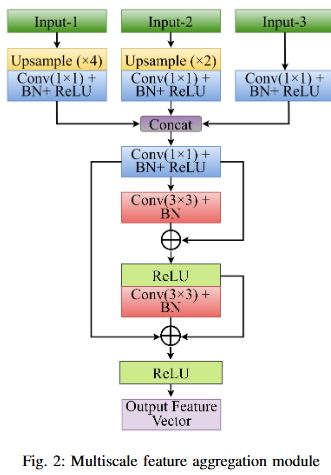
Abstract
Automated colonoscopic polyp segmentation, vital for early colorectal cancer detection, has notably advanced with deep learning. However, reliance on operator expertise and challenges posed by varied polyp sizes during training, along with limitations in representing long-range dependencies, warrant further refinement in the field. In this paper, we propose a novel transformer-based text guided architecture called TGA-TransNet to address these challenges. Particularly, the TGA-TransNet model incorporates size-related and polyp number-related information through text attention during training. Additionally, Transformer layers are utilized to extract global contexts. Our early experimental results on the popular Kvasir-SEG dataset indicate that the proposed integration of text embeddings and Transformer enhances the overall segmentation performance of the model, surpassing its state-of-the-art counter-part. Feature map analysis also suggests that our proposed TGA-TransNet demonstrates strong generalization capabilities across the variable-sized polyps.
BibTeX
Click to copy@INPROCEEDINGS{10561827,
author={Ovi, Tareque Bashar and Bashree, Nomaiya and Nyeem, Hussain and Wahed, Md Abdul and Alam, Ayat Subah and Rhythm, Faiaz Hasanuzzaman},
booktitle={2024 3rd International Conference on Advancement in Electrical and Electronic Engineering (ICAEEE)},
title={A Transformer-based Text-Guided Approach for Improved Colonoscopic Polyp Segmentation},
year={2024},
volume={},
number={},
pages={1-6},
keywords={Training;Deep learning;Image segmentation;Image coding;Colonic polyps;Predictive models;Transformers;deep learning;medical image segmentation;contextual information;text guided attention;transformer},
doi={10.1109/ICAEEE62219.2024.10561827}}
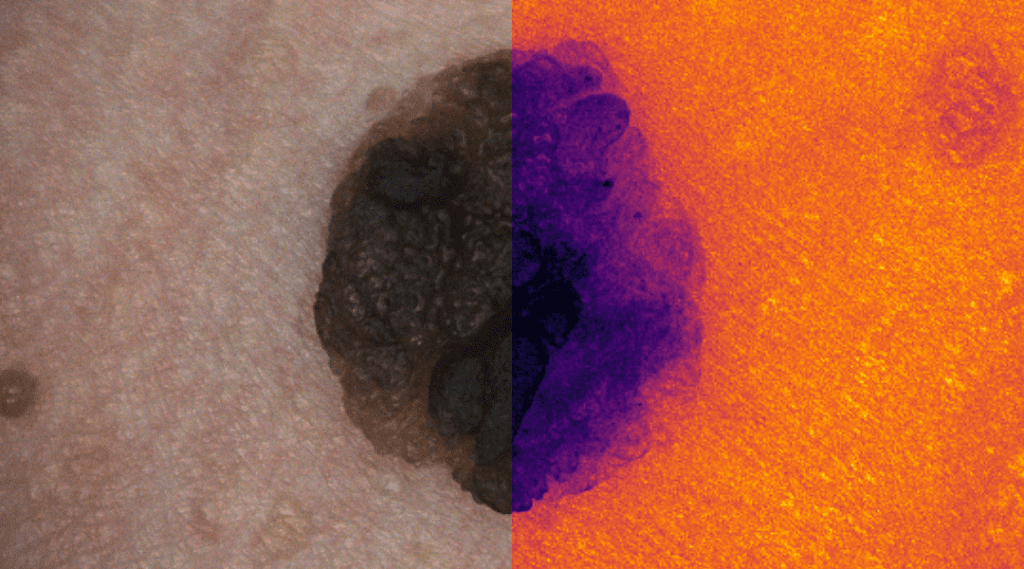
Skin Cancer Detection
Deep learning for skin cancer relies on segmentation and classification.
This research area focuses on developing advanced deep learning frameworks for the automated detection, segmentation, and classification of skin lesions from dermoscopic and clinical images. By leveraging convolutional neural networks (CNNs) and transformer-based architectures, the goal is to differentiate malignant melanomas from benign skin conditions with high diagnostic accuracy. The research integrates explainable AI techniques to enhance clinical trust and decision support, ultimately contributing to early detection and improved patient outcomes in dermatological diagnostics.
Selected Research Projects
Objective
No objective provided.
Key Findings
No findings listed yet.
Photos
No photos added.
Ongoing Research Projects
Objective
This study evaluates the robustness of leading segmentation models against advanced adversarial attacks in the context of polyp and skin lesion identification.
Key Findings
- We benchmark the robustness of leading segmentation models under strong adversarial attacks, systematically identifying their failure modes in the context of polyp and skin lesion tasks.
- The first systematic study of bi-level nested architectures under adversarial threat, clarifying how multiscale fusion and cross-level aggregation influence resilience.
- Evidence that global feature representations mitigate adversarial perturbations, with distilled design guidelines for more robust medical segmentation systems.
Photos
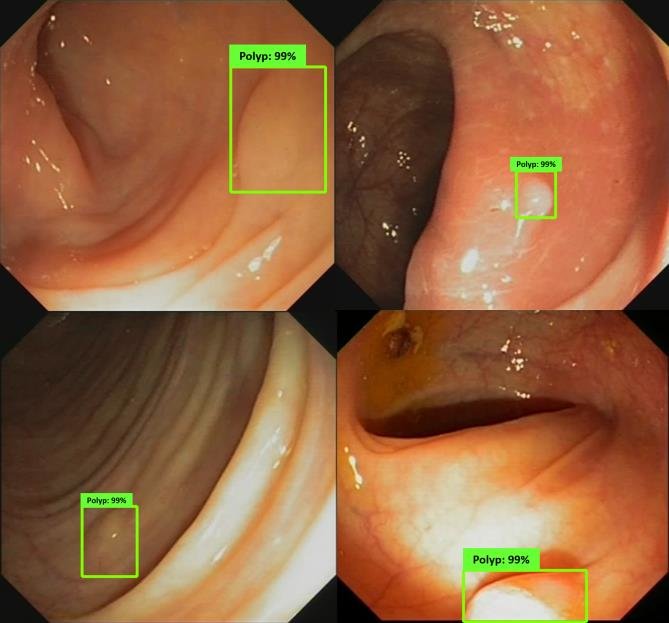
Colonoscopic Polyp Detetction
An AI "second eye" for real-time polyp detection during colonoscopy.
Research in colonoscopic polyp detection aims to design real-time AI-assisted systems for the early identification of colorectal polyps during colonoscopy. Using deep convolutional and attention-based segmentation models, the objective is to improve the sensitivity and precision of polyp localization under varying illumination, motion, and texture conditions. The developed models function as a “second observer” for endoscopists, assisting in diagnostic decisions and reducing the rate of missed polyps, thereby enhancing colorectal cancer prevention.
Selected Research Projects
No selected projects in this area.
Ongoing Research Projects
Objective
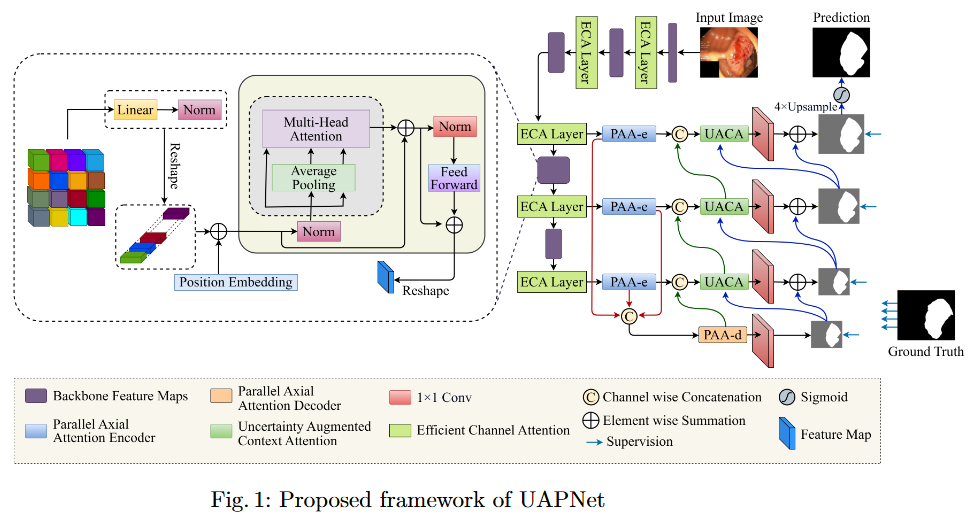
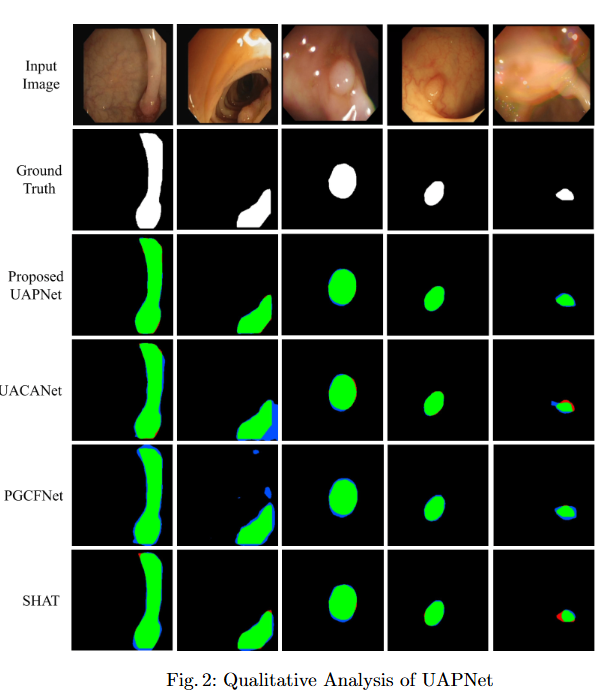
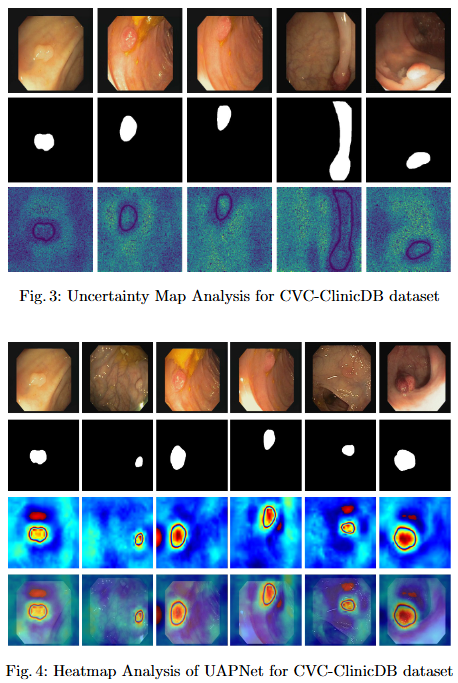
To mitigate feature redundancy in deep networks, we integrate the Efficient Channel Attention (ECA) module with the PVT encoder and augment it with an Uncertainty Augmented Context Attention (UACA) mechanism.
Key Findings
- PVT with Uncertainty-Augmented Context Attention. We integrate the PVT encoder with an uncertainty-augmented context attention mechanism, enabling the model to focus explicitly on ambiguous boundary regions. This mechanism improves the accuracy of boundary delineation by using uncertainty as a guiding signal.
- Integrating ECA with PVT Based Encoder. We incorporate ECA after each PVT stage, which refines feature representations while maintaining the original dimensionality. This enhancement improves the model’s discriminative power without introducing additional computational overhead.
- Explainability for Clinical Reliability. To ensure transparency and clinical applicability, we apply explainable AI (XAI) techniques, including uncertainty maps and heatmaps for foreground and edge regions. These methods improve interpretability and provide clinicians with more reliable decision making tools by highlighting areas of uncertainty and focus during polyp segmentation.
Photos
Objective
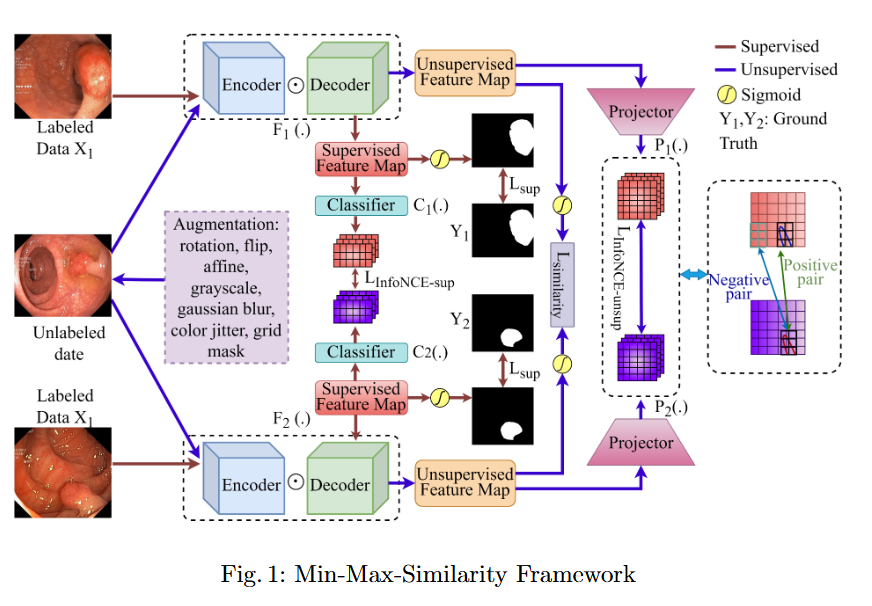
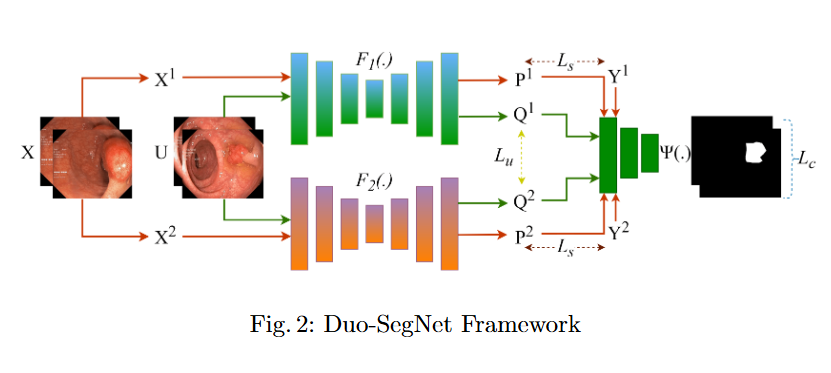
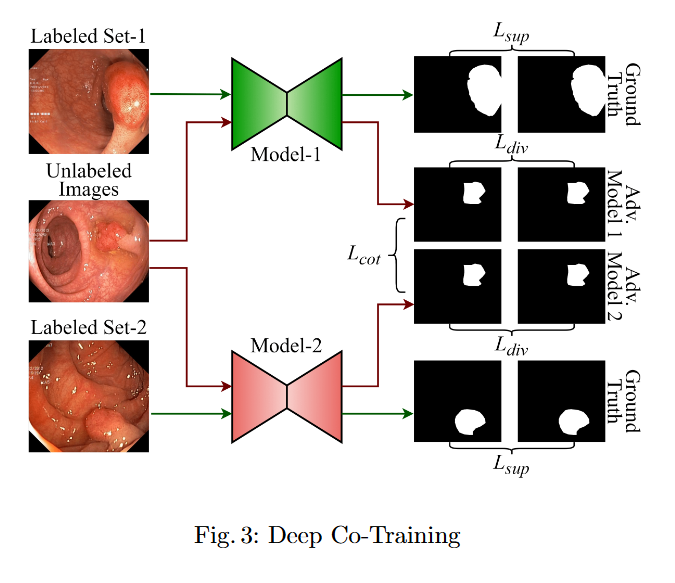
In this study, we systematically investigate how established supervised segmentation models perform within three representative semi-supervised frameworks: Min-Max Similarity, Duo-SegNet, and Deep Co-Training.
Key Findings
- We represent a comprehensive evaluation of multiple supervised segmentation architectures in semi-supervised learning scenarios.
- We identify model characteristics that enable effective utilization of unlabeled data within semi-supervised frameworks.
- We provide empirical insights that inform the design of more better and dependable semi-supervised polyp segmentation methods, thereby reducing annotation requirements while maintaining high segmentation accuracy.
Photos
Objective
This study evaluates the robustness of leading segmentation models against advanced adversarial attacks in the context of polyp and skin lesion identification.
Key Findings
- We benchmark the robustness of leading segmentation models under strong adversarial attacks, systematically identifying their failure modes in the context of polyp and skin lesion tasks.
- The first systematic study of bi-level nested architectures under adversarial threat, clarifying how multiscale fusion and cross-level aggregation influence resilience.
- Evidence that global feature representations mitigate adversarial perturbations, with distilled design guidelines for more robust medical segmentation systems.
Photos

Medical Image Analysis
Imaging, segmentation, and diagnostics for medical data.
This area encompasses a broad spectrum of imaging modalities—MRI, CT, X-ray, and endoscopic imaging—to develop computational tools for segmentation, registration, and diagnostic interpretation. Research focuses on integrating multimodal image features with deep learning to improve disease localization and treatment planning. The emphasis is on constructing efficient, explainable, and data-driven models that support clinical workflows, thereby bridging the gap between medical imaging research and practical healthcare applications.
Selected Research Projects
Objective
No objective provided.
Key Findings
No findings listed yet.
Photos



Objective
No objective provided.
Key Findings
No findings listed yet.
Photos




Objective
No objective provided.
Key Findings
No findings listed yet.
Photos



Ongoing Research Projects
No ongoing projects in this area.

Computer Vision
Algorithms and systems for visual understanding and perception.
Computer vision research at the lab explores the development of algorithms and systems capable of perceiving and understanding visual data in real-world environments. Topics include object detection, scene understanding, tracking, and visual reasoning. By integrating attention mechanisms, transformers, and graph neural networks, this work advances autonomous perception systems across domains such as healthcare, robotics, and intelligent surveillance, pushing the boundaries of visual cognition and artificial intelligence.
Selected Research Projects
Objective
No objective provided.
Key Findings
No findings listed yet.
Photos



Ongoing Research Projects
No ongoing projects in this area.

Remote Sensing
Earth observation and geospatial analysis from aerial and satellite imagery.
This research investigates the use of aerial and satellite imagery for earth observation and geospatial analysis. Deep neural networks are employed for tasks such as land cover classification, road and building extraction, and environmental monitoring. The goal is to develop accurate and scalable models capable of processing high-resolution satellite data, thereby supporting urban planning, disaster management, and sustainable resource monitoring with precision-driven remote sensing analytics.
Selected Research Projects
Objective
To develop SE-U2Net, a robust and efficient deep learning framework for road extraction from very high-resolution (VHR) satellite imagery by integrating the U²-Net architecture with Squeeze-and-Excitation (SE) modules, enabling selective channel refinement and multi-scale contextual feature learning without reliance on pre-trained backbones.
Key Findings
- Architectural Innovation: Integration of Squeeze-and-Excitation (SE) blocks for dynamic channel feature recalibration.
- Performance Improvement: Enhanced discrimination of minor roads through learned channel interdependencies.
- Computational Efficiency: Parameter-efficient attention mechanism achieved without complex spatial operations.
Photos



Objective
To develop DoubleUNet++, an enhanced road extraction model that improves the contextual understanding and spatial precision of remote sensing image segmentation by integrating a Channel-Aware Gated Attention (CAGA) mechanism and Squeeze-and-Excitation (SE) layers into the DoubleUNet architecture.
Key Findings
- We propose Channel-Aware Gated Attention (CAGA) mechanism which has been integrated into skip connections of the secondary U-Net, enabling dynamic spatial feature weighting to emphasize critical road regions while suppressing irrelevant details
- A squeeze-and-excitation (SE) layer preceding the final activation in the attention pathway, adaptively recalibrating channel-wise feature responses to enhance discriminative power
Photos




Ongoing Research Projects
No ongoing projects in this area.

Computational Biology and Bioinformatics
Data-driven modeling and analysis of biological systems.
The lab’s research in computational biology and bioinformatics applies machine learning and statistical modeling to the analysis of complex biological data. Efforts include protein structure prediction, gene expression analysis, and biological network modeling. Through the integration of omics data and imaging modalities, this area aims to uncover biological patterns and enhance understanding of disease mechanisms, ultimately contributing to precision medicine and drug discovery initiatives.
Selected Research Projects
No selected projects in this area.
Ongoing Research Projects
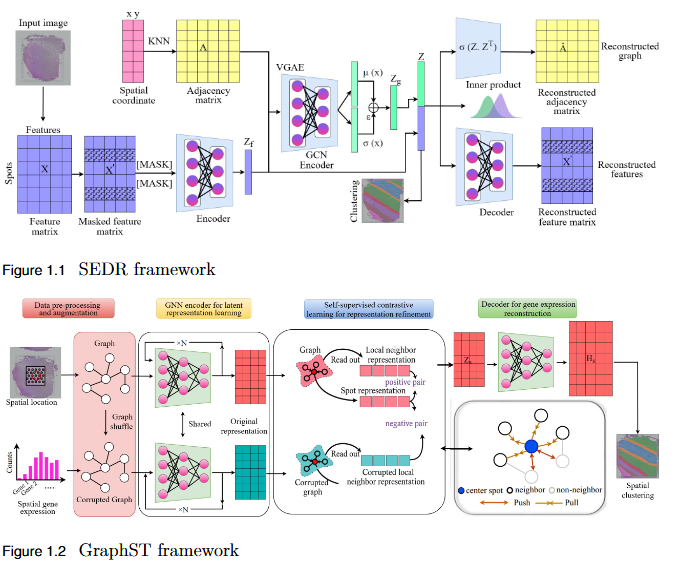
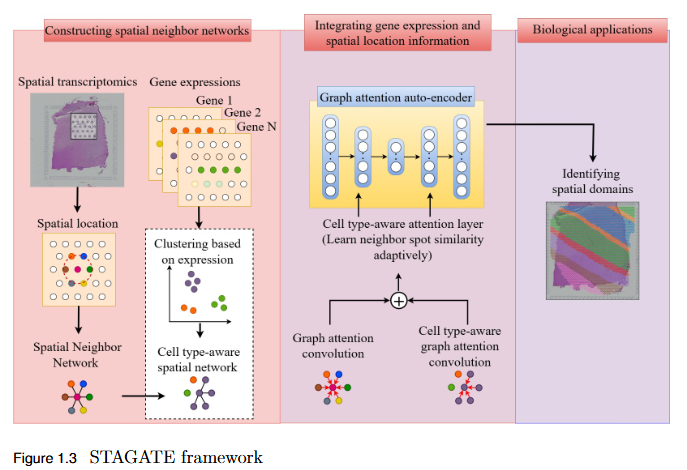
Objective
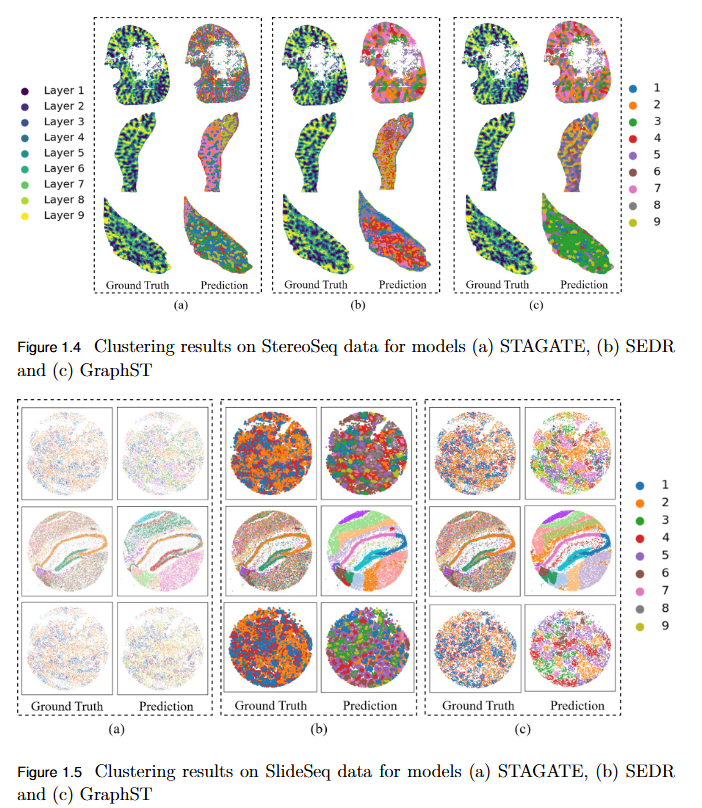
To systematically evaluate the performance and scalability of leading GNN-based spatial transcriptomics (ST) models—STAGATE, GraphST, and SEDR—on high-density next-generation datasets such as Stereo-seq and Slide-seq, and to identify their limitations in modeling complex spatial structures.
Key Findings
- Performance Evaluation: Comprehensive assessment using ARI, AMI, NMI, and HOMO metrics revealed modest and variable clustering performance across high-density datasets.
- - SEDR achieved the highest scores on Slide-seq datasets, but overall accuracy remained low.
- - Stereo-seq results were inconsistent, with no single model consistently outperforming others.
- Scalability Limitation: Existing GNN-based ST models struggle to generalize to high-resolution data, indicating poor scalability to large, complex tissue structures.
- Benchmark Establishment: This study provides a performance baseline for evaluating future models on next-generation ST platforms.
- Research Implication: Highlights the urgent need for developing new computational frameworks that are more robust, adaptive, and scalable for analyzing dense spatial transcriptomics data.
Photos
Objective
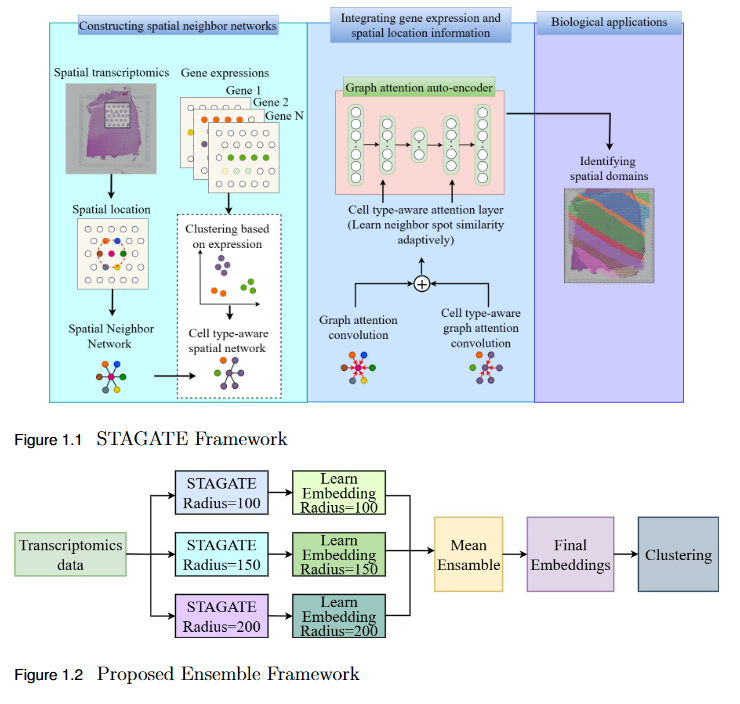
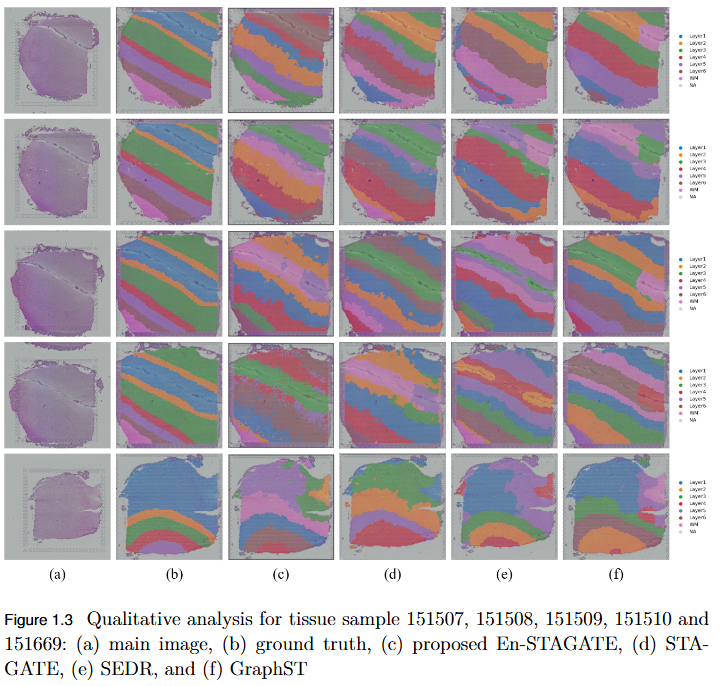
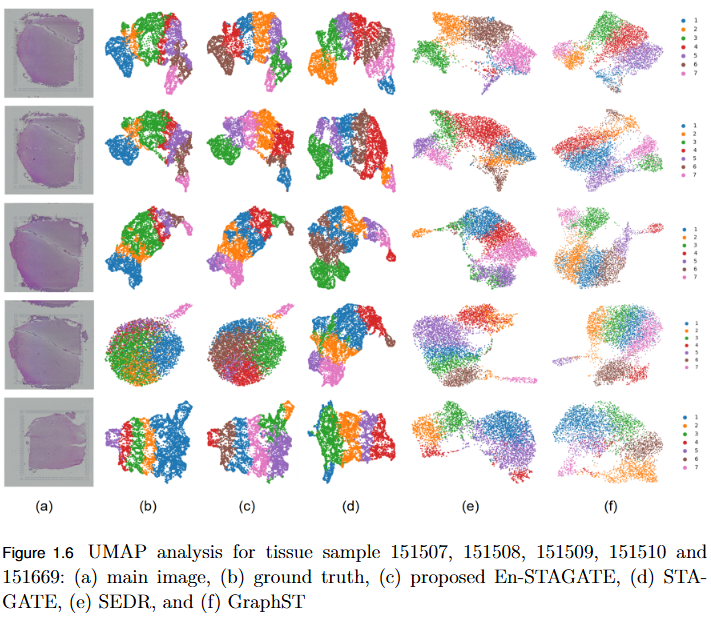
To overcome the limitation of fixed spatial neighborhood scales in existing spatial transcriptomics (ST) models by developing En-STAGATE, an ensemble framework that integrates multiple spatial graphs across varying neighborhood radii to capture both local and global tissue structures for improved domain identification.
Key Findings
- Multi-Scale Ensemble Design: En-STAGATE constructs and fuses representations from multiple spatial graphs at different neighborhood radii, effectively modeling both fine-grained cellular interactions and large-scale tissue organization.
- Enhanced Biological Interpretability: The multi-scale integration provides a more comprehensive and biologically meaningful latent representation of tissues.
- Empirical Validation: Achieved competitive or superior performance across 14 benchmark datasets, outperforming STAGATE and other leading methods in several key cases.
- Improved Domain Delineation: Notably higher Adjusted Rand Index (ARI) scores on datasets such as 151674, 151675, 151676, and Mouse, demonstrating robustness in complex tissue structures.
- Key Insight: Highlights that a single neighborhood scale is insufficient for spatial domain identification, establishing multi-scale analysis as a critical paradigm in spatial transcriptomics.
Photos
Objective
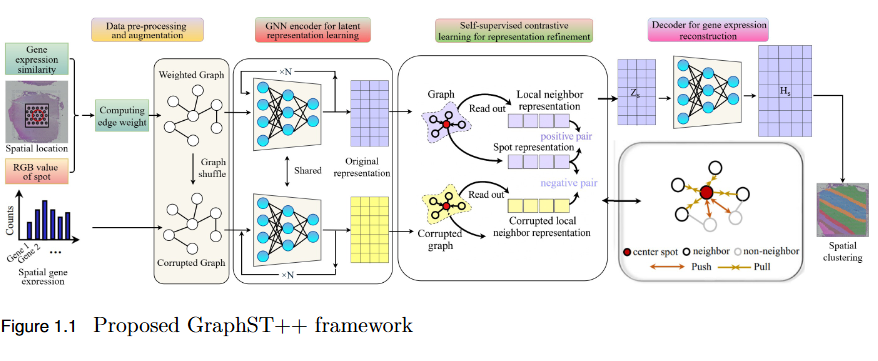
To address the over-smoothing and spatial bias in current spatial transcriptomics (ST) models by developing GraphST++, a multi-modal framework that integrates spatial, gene expression, and histological features for constructing biologically faithful tissue graphs.
Key Findings
- Multi-Modal Graph Construction: GraphST++ defines inter-spot relationships using a composite score combining spatial distance, gene expression similarity, and histological morphology, reducing over-reliance on proximity-based connections.
- Improved Tissue Representation: The proposed approach captures complex biological interactions and produces a more accurate reflection of the tissue microenvironment.
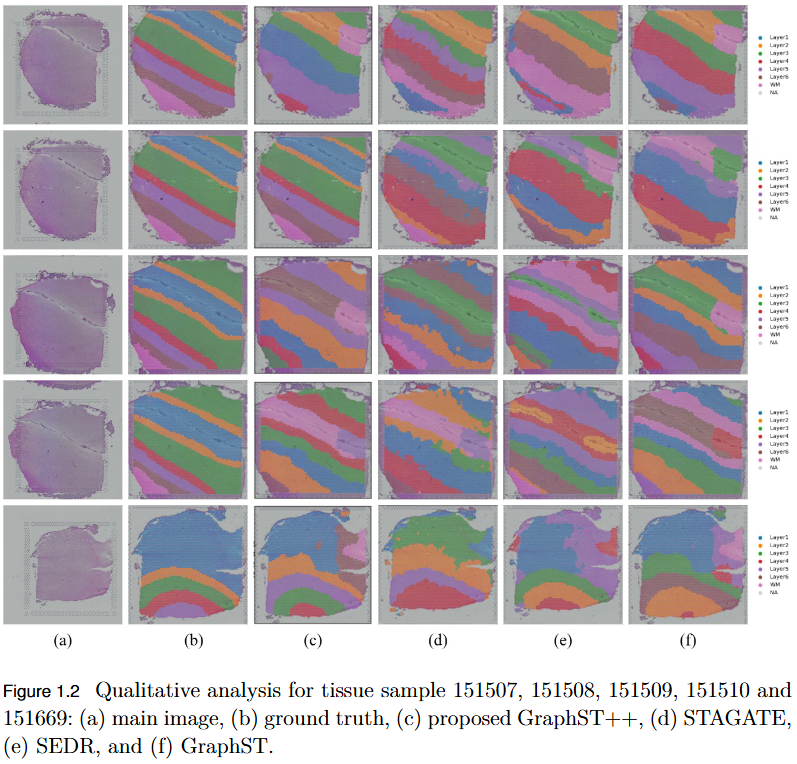
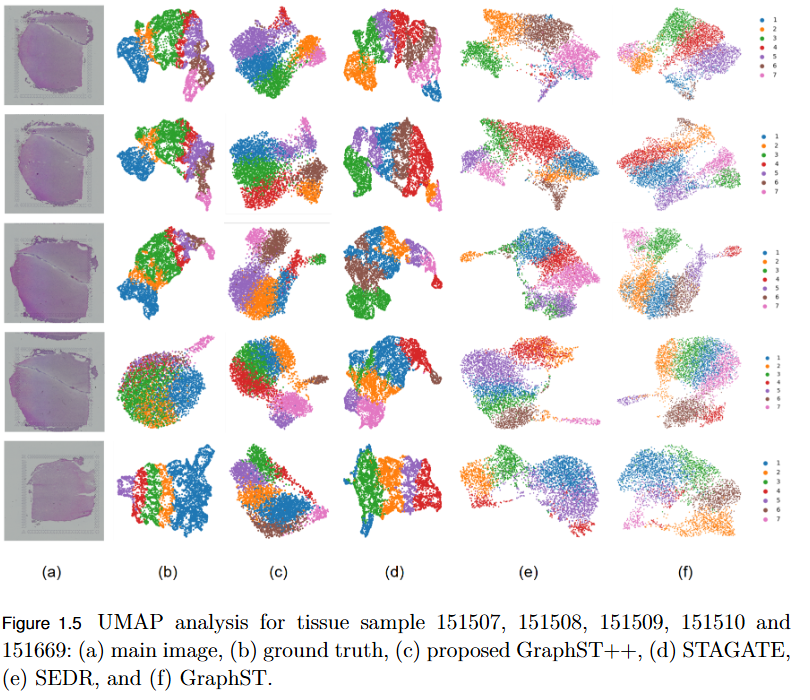
- Superior Performance: Demonstrated higher Adjusted Rand Index (ARI) compared to leading models, including GraphST, across multiple benchmark datasets such as human breast cancer (BRCA) and human brain tissues.
- Robustness Across Datasets: While not universally dominant, GraphST++ consistently benefits from morphological integration, yielding more stable and biologically interpretable clustering results.
- Broader Implications: Highlights the importance of multi-modal fusion in advancing spatial transcriptomics analysis and enabling more precise tissue domain identification.
Photos
Objective
To overcome the limitation of existing graph-based spatial transcriptomics (ST) models in capturing long-range spatial dependencies by developing STAGATE++, a hybrid GAT–Transformer architecture that jointly learns local and global tissue representations.
Key Findings
- Hybrid Architecture Advantage: STAGATE++ integrates a Graph Attention Network (GAT) for precise local feature extraction with a Transformer encoder for modeling long-range spatial dependencies.
- - Enhanced Spatial Awareness: The model constructs a holistic and spatially coherent tissue representation, improving understanding of tissue organization.
- Empirical Superiority: Demonstrated state-of-the-art performance across 14 benchmark datasets, consistently outperforming existing methods in spatial domain identification.
- Validation Metric: Significant improvements in Adjusted Rand Index (ARI) confirm the importance of global context modeling in tissue delineation.
- Future Extensions: Plans include incorporating histopathological image features, applying self-supervised training for better generalization, and integrating GNN interpretability techniques for biological insight and clinical transparency.
Photos
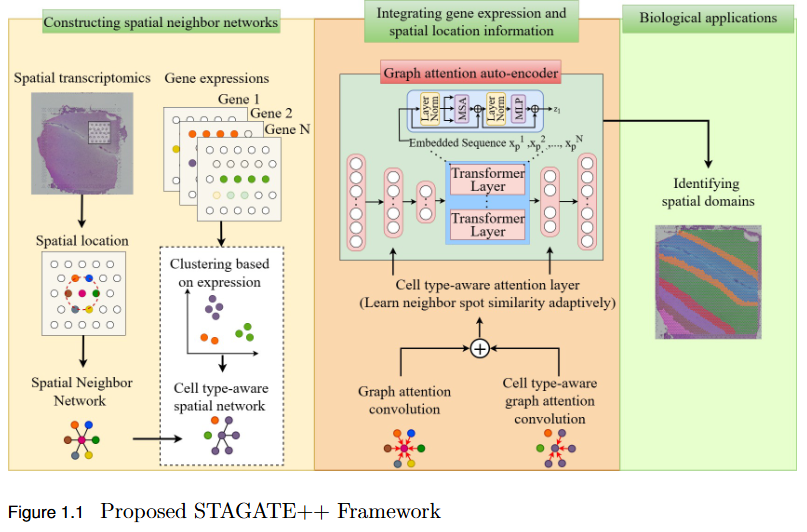
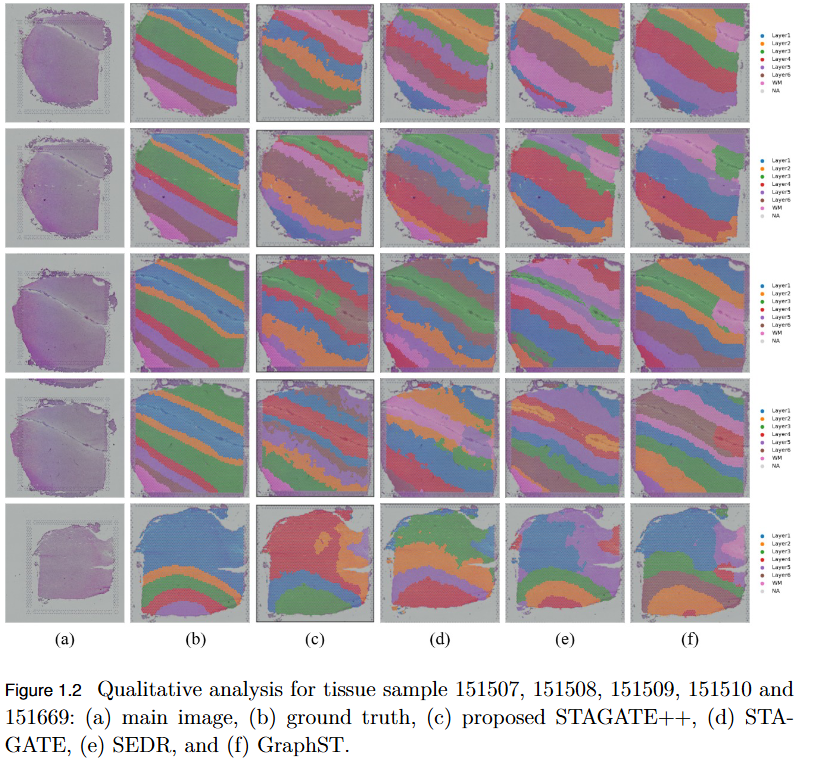
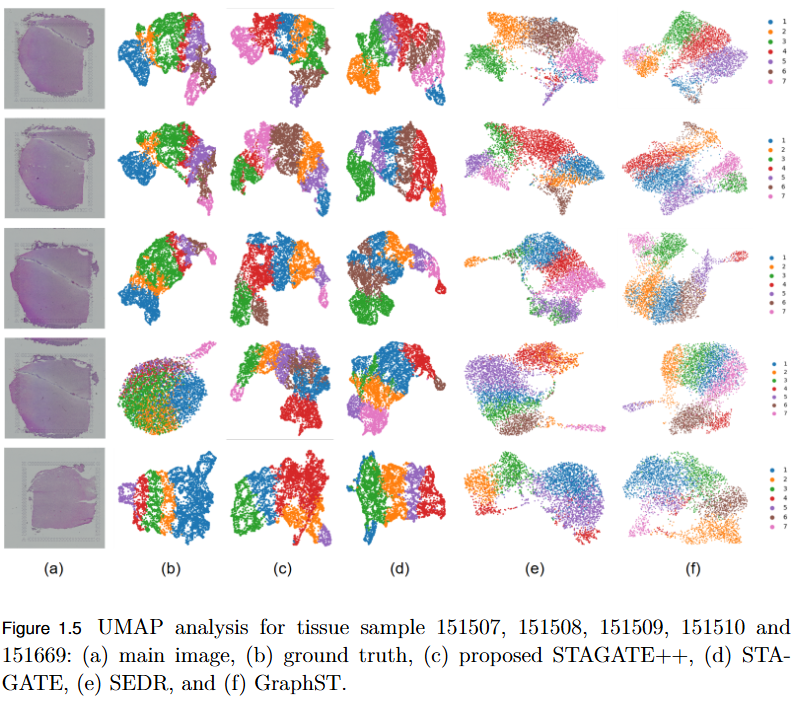

Image Processing
Signal and image enhancement, restoration, and transformation techniques.
This foundational area focuses on the theoretical and practical aspects of image enhancement, restoration, compression, and transformation. Research includes spatial and frequency-domain techniques for noise reduction, contrast enhancement, and feature extraction. By combining classical methods with modern deep learning frameworks, the objective is to achieve efficient, high-quality image processing pipelines that serve as the backbone for advanced applications in computer vision, medical imaging, and multimedia analysis.
Selected Research Projects
Objective
No objective provided.
Key Findings
No findings listed yet.
Photos


Ongoing Research Projects
No ongoing projects in this area.
